A new paper led by DBEI faculty member Mingyao Li, PhD, introduces smart spatial omics (S2-omics) — an automated workflow designed to make spatial omics experiments more rigorous, efficient, and reproducible.
The paper “Smart spatial omics (S2-omics) optimizes region of interest selection to capture molecular heterogeneity in diverse tissues” introduces a powerful new workflow designed to improve the rigor, efficiency, and impact of spatial omics experiments.
In spatial omics, researchers profile molecular information (e.g., transcripts or proteins) across tissues while preserving the spatial context — a major advance in understanding tissue architecture, disease microenvironments, and cellular interactions.
Yet because spatial omics assays are expensive and only cover small portions of large tissue sections, selecting which regions to profile (regions of interest, or ROIs) is usually done manually — a subjective, inconsistent, and often non-reproducible process.
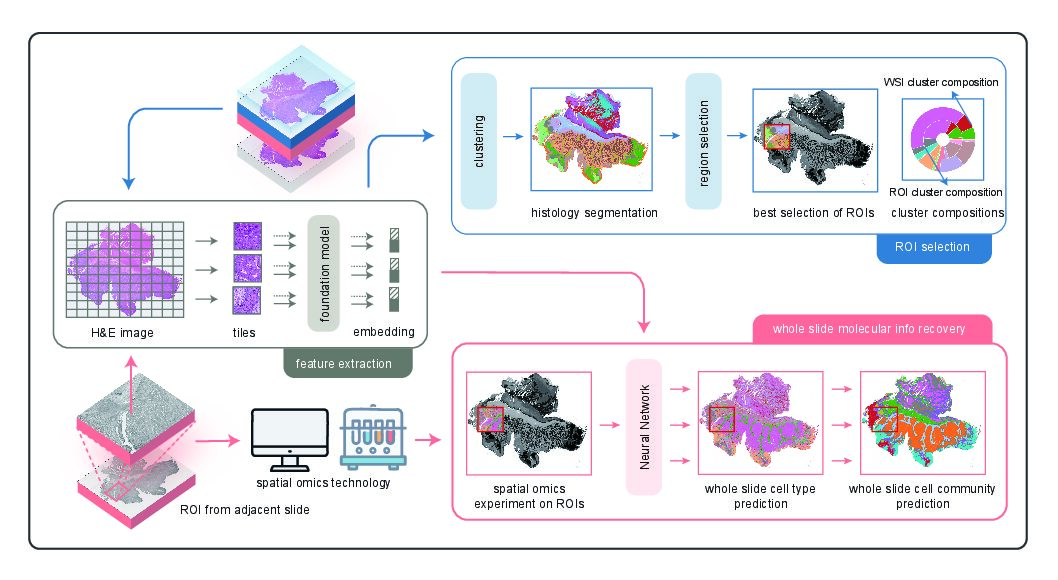
S2-omics solves this bottleneck by using only standard H&E histology images to automatically select ROIs that maximize molecular information content. The method extracts image features with pre-trained pathology “foundation models,” clusters the tissue into distinct histological regions, then selects ROIs using a quantitative “ROI score” to ensure representative sampling of tissue heterogeneity.
Benchmarking across multiple tissue types (breast, colon, kidney, liver, stomach) and spatial omics platforms (e.g., Xenium, Visium HD, CosMx), the paper showed that S2-omics often matches or outperforms manual expert selection. Importantly, S2-omics–chosen ROIs captured critical structures, including tumors and tertiary lymphoid structures, and enabled accurate whole-slide cell-type and community predictions from limited sampling.
By standardizing and automating ROI selection, S2-omics reduces subjectivity, increases reproducibility, and helps ensure that costly spatial omics efforts yield maximal biological insight. This represents a new foundational tool that can make spatial omics studies more efficient, scalable, and robust.