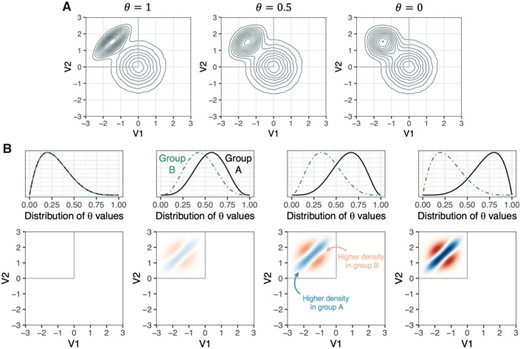
A great deal of neuroimaging research focuses on voxel-wise analysis or segmentation of damaged tissue, yet many diseases are characterized by diffuse or non-regional neuropathology. In simple cases, these processes can be quantified using summary statistics of voxel intensities. However, the manifestation of a disease process in imaging data is often unknown, or appears as a complex and nonlinear relationship between the voxel intensities on various modalities. When the relevant pattern is unknown, summary statistics are often unable to capture differences between disease groups, and their use may encourage post hoc searches for the optimal summary measure. In this study, we introduce the multi-modal density testing (MMDT) framework for the naive discovery of group differences in voxel intensity profiles. MMDT operationalizes multi-modal magnetic resonance imaging (MRI) data as multivariate subject-level densities of voxel intensities and utilizes kernel density estimation to develop a local two-sample test for individual points within the density space. Through simulations, we show that this method controls type I error and recovers relevant differences when applied to a specified point. Additionally, we demonstrate the ability to maintain power while controlling the family-wise error rate and false discovery rate when applying the test over a grid of points within the density space. Finally, we apply this method to a study of subjects with either multiple sclerosis (MS) or conditions that tend to mimic MS on MRI, and find significant differences between the two groups in their voxel intensity profiles within the thalamus. Dworkin et al. (2019)